My genome: Let me show you it
tl;dr: download Rob’s source code
In October 2015 I signed up as a beta tester for Arivale, a Seattle-based “scientific wellness” company. The service is something like nutritional-coach-meets-quantified-self.
In their words:
Our systems approach gathers, connects, and analyzes your data to create a complete picture of you.
And that it does.
Once a month I have a chat with a nutritional coach about my current diet, life stresses, and exercise habits. Over the course of a year they take multiple blood samples and plot an extensive panel of blood chemistry trends over time. They collect multiple saliva samples, measuring cortisol at four points throughout the day. They perform a gut microbiome sequencing (gross, yet fascinating!) to measure the impact of diet on microbial population diversity. They supply a Fitbit to track steps, sleep, and heart rate. They take a DNA sample and run a SNP panel looking for several variations linked to nutrition and exercise.
And last (but certainly not least), they perform whole genome sequencing. This sets it solidly apart from services like 23andMe that can only detect specific SNPs. While the whole genome is specifically excluded from the coaching process, it is used (with consent) as a basis for further genomic study.
Most importantly: Arivale provides a copy of the data, including a VCF and the raw reads.

After anxiously waiting for several months, I finally received an encrypted hard drive containing a VCF file and an aligned BAM file. Tech specs for the reads:
- Ran on an Illumina HiSeq X Ten
- 106 GB of compressed BAM data
- 150 bp paired reads
- Just under 600 million reads total
- 30x average coverage
- Uses hs37d5 for a reference
- FastQC indicates that the read quality is quite good:
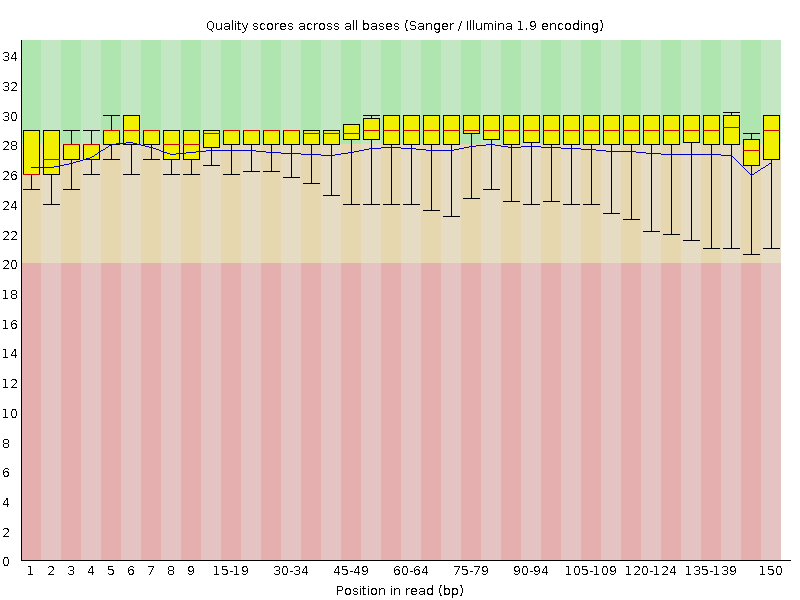
The VCF calls about 4.5 million variants, including standard rsids. The longest called deletion is 231 bases, and the longest insertion is 524 bases.
But what does it all mean?
That, my friends, is an ongoing and evolving field of study.
The human genome itself was first sequenced in 2003 (coincidentally, just after I moved to Seattle). But 13 years later, we do not yet have a simple database where you can look up “what a gene does” or “what a genetic variation means”.
The current state of the art includes databases like dbSNP and dbVar and clinVar that attempt to tie genetic samples together with studies of specific phenotypes and conditions. It’s new science, and still tough going.
It’s not clear that we will ever have a database that tells us “what this gene does”, because life is clearly much more complex than that. DNA is Layer 1 of the stack that runs this program called life. Epigenetics and microbiota and environment and poor life choices clearly have a significant impact on the health of any given organism.
And yet, DNA provides the ground rules of what any organism could aspire to. Cats beget cats. Plants beget plants. Bacteria beget bacteria. People beget people (who host a colony of bacteria at least as big as they are).
Your DNA is not your body, but it does set the parameters for what can be made with locally available materials.
As a hacker, I’d like to help document and debug Layer 1. Now that I have a copy of my source code, I intend to share the code review process with you.
Responsible disclosure
There are already many online sources of human genetic data available for analysis (see 1000 Genomes, the NCBI Sequence Read Archive, the European Nucleotide Archive, etc.) Researchers benefit from large and factually complete databases that make it possible to perform genome-wide association studies that can link genetic traits to phenotype and disease risk in a way that would not otherwise be possible.
But our genetic data tells possibly the most intimate story about ourselves, including our ancestral background, inherited disease risks, and direct family relations. Data mining can turn up many unexpected patterns. Some happy, some not so happy.
For that reason, public genetic databases take personal privacy (and HIPAA compliance) seriously. And I’m sure they don’t want to be sued.
Ideally I’d like my genetic data to be studied as widely and thoroughly as possible. To alleviate all possible privacy concerns, I hereby release my own genome under Creative Commons CC-BY-SA. You may reuse or remix my genetic data on a non-commercial basis any way you like. Please share your findings!
And I’d appreciate an introduction to any evil clones you might produce. Just don’t forget to credit the original author. (Spoiler alert: they’re all evil.)
My data is up on the SRA with ID SRR3990320. It’s also referenced by BioProject PRJNA335906. To download the data, it’s best to use sratools or ascp; a slow and often unreliable ftp link should also be up shortly. The BAM is 106 GB.
While it’s apparent that people of European descent are already overrepresented in modern genomics, nobody else’s genome is mine to give. I expect this gap to close sharply as the cost of sequencing continues to plummet and it becomes a standard test covered by insurance. In the meantime, I hope one more white dude’s data is useful to somebody.
Curious about how your genes determine your eye color? Look Into My Eyes.